Human Coronavirus 2019-nCov Spike
SARS-CoV-2 Spike
Spike (S) is a structural glycoprotein expressed at the SARS-CoV-2 (2019-nCoV) surface and is a critical determinant of the viral host and tissue tropism. SARS-CoV-2 S mediates the virus entry into the target cells upon ACE2 receptor binding [1, 2], and is thus a potential therapeutic drug-target [3, 4]. Elevated Anti-SARS-CoV-2 S antibody titers are detected in COVID-19 patients’ sera [3, 4], rising approximately ten days after symptoms onset [3]. These observations make SARS-CoV-2 S an attractive tool for early diagnosis [3-5]. Spike antigenicity is also highlighted by the observation of Spike-specific CD4+ and CD8+ T cells in blood samples from recovered COVID-19 patients [5]. As a consequence, SARS-CoV-2 S has been the main target for prophylactic vaccination strategies [6-8].
Spike protein overview
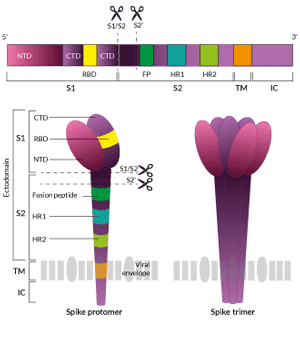
Most of the current knowledge about SARS-CoV-2 S is based on analogies with findings on the previously identified SARS-CoV S. Clove-shaped trimers of Spike proteins form large surface protrusions that give the coronaviruses the appearance of having a crown. Each Spike protomer contains three segments: a large ectodomain, a transmembrane anchor (TM), and a short intracellular tail (IC) [1, 2, 9]. The Spike ectodomain contains three critical elements:
- the S1 subunit contains an N-terminal (S1-NTD) and a C-terminal (S1-CTD) subdomains. The S1 "closed" conformation exerts a physical constraint on the S2 subunit until specific proteases cleave the S1/S2 and S2' sites [9].
- the RBD (receptor binding domain) is located in the S1-CTD region and is buried in the inner S1 head-trimer. The S1 "open" conformation is expected to be necessary for binding to the ACE2 receptor at the surface of host target cells [9, 10].
- the S2 subunit forms a trimeric stalk. It contains a fusion peptide (FP) and two heptad repeats (HR1 and HR2), which operate the fusion of viral and host membranes [9, 10].
Two cleavage sites at the S1 and S2 boundary (S1/S2) and in the S2 domain (S2’) play an essential role in the viral entry into host target cells [9-12].
Virus fusion with the host cell membrane
The Spike protein exists in two structurally distinct conformations: pre-fusion and post-fusion. In its pre-fusion state, Spike is a "closed" trimer and RBDs are buried in the inner S1 head-trimer, at the interface between each protomer [2]. This "closed" conformation exerts a physical constraint on the S2 subunit until specific proteases cleave the S1/S2 and S2' sites [3]. The exact mechanisms driving the opening of an S1-CTD domain and the subsequent exposition of RBD so that it can bind the ACE2 receptor are not elucidated yet. It has been proposed that the S protein is cleaved into S1 and S2 subunits by proteases, including furin, the host surface-associated transmembrane protease serine 2 (TMPRSS2), and the endocytic cathepsin L [9-12]. S1 binds to ACE2 through its RBD, and S2 is further cleaved and activated by TMPRSS2 and/or cathepsin L [9, 10]. Together these actions result in host-viral membrane fusion and release of the viral RNA genome into the host cell cytoplasm.
SARS-CoV-2 Spike variants
Early in the pandemic, the D614G amino acid mutation was identified within the spike protein and has rapidly become the dominant variant around the world. The D614G mutation is located at the C-terminus of the S1 domain, near the furin cleavage site [13].
There is currently no scientific consensus on a positive selection for the G614 variant where it would associate with higher infectivity and transmissibility in humans [14]. Importantly, antibodies raised against SARS-CoV-2 D614 potently neutralize infection with its G614 counterpart in a hamster model [15], suggesting that the D614G mutation may not reduce the protective ability of vaccines in clinical trials.
SARS-CoV-2 Spike in vaccination strategies
The previous SARS-CoV and MERS-CoV outbreaks have prompted the search for protective vaccines. Most vaccine candidates for COVID-19 aim to induce neutralizing antibodies against Spike, preventing uptake via the human ACE2 receptor and thereby blocking infection [6-8].
Three main vaccination strategies rely on SARS-CoV-2 S antigenicity and use either DNA, RNA, or protein platforms [8]. Below are three representative examples:
- The INO-4800 vaccine developed by Inovio consists of a DNA plasmid encoding Spike that is delivered using the CELLECTRA® 2000 electroporator (NCT04336410; phase I/II as of September 2020).
- The mRNA-1273 vaccine developed by Moderna consists of a non-replicating mRNA encoding the full-length prefusion stabilized Spike, mixed proprietary lipid nanoparticles, delivered by intramuscular injection (NCT04283461; phase II as of September 2020).
- The NVX-CoV2373 vaccine developed by Novavax consists of a recombinant Spike protein in its pre-fusion state incorporated into a nanoparticle and delivered by intramuscular injection along with a proprietary adjuvant (NCT04368988; Phase I/II as of September 2020).
Depending on your applications, InvivoGen offers:
➤ Plasmids harboring the genes encoding SARS-CoV-2 Spike. These ready-to-go vectors have been designed for mammalian cell expression or protein production and secretion.
➤ Fusion proteins consisting of Spike fragments with a C-terminal poly-histidine or human IgG1 Fc tag.
➤ Antibodies targeting the SARS-CoV-2 Spike RBD. These antibodies are available with different immunoglobulin isotypes, either native or engineered.
![]() Read our reviews on COVID-19:
Read our reviews on COVID-19:
➤ The infection cycle of SARS-CoV-2
➤ Treatment with repurposed drugs
➤ Predicted host immune responses to SARS-CoV-2
➤ Vaccine development
➤ Protective immunity & Re-infection
Products
| Spike (S), ectodomain, S1, and RBD Coding Sequences | Genes encoding SARS-CoV-2 Spike in vectors designed for mammalian cell expression or secretion |
| SARS-CoV-2 Spike RBD Proteins | Recombinant fusion proteins produced in CHO cells |
| SARS-CoV-2 Spike S1 Proteins | Recombinant fusion proteins produced in CHO or HEK293 cells |
| Anti-SARS-CoV Spike mAb (clone CR3022), human & mouse isotypes | Anti-Spike RBD antibody - Human & Mouse isotypes |
References
1. Li F., 2016. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 3:237-261.
2. Li F. et al., 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 309:1864-1868.
3. Wu Y. et al., 2020. A non-competing pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 368(6496):1274-1278.
4. To K. K-K. et al., 2020. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational study. The Lancet Infectious Diseases. 20(5):565-574.
5. Grifoni A et al., 2020. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 181(1489-1501).
6. Wang N. et al., 2020. Subunit vaccines against emerging pathogenic human coronaviruses. Front. Microbiol. 11:298. DOI: 10.3389/fmicb.2020.00298.
7. Padron-Regalado E., 2020. Vaccines for SARS-CoV-2: Lessons from other coronavirus strains. Infect. Dis. Ther. DOI: 10.1007/s40121-020-00300-x.<
8. Funk C.D et al., 2020.A Snapshot of the global race for vaccines targeting SARS-CoV-2 and the COVID-19 pandemic. Front. Pharmacol. 11:937.
9. Walls A.C. et al., 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 181(2):281-292.e6.
10. Hoffmann M. et al., 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 181:1-16.
11. Hoffmann M. et al., 2020. A multibasic cleavage site in the Spike protein of SARS-CoV-2 is essential for infection of human lung cells. Molecular Cell. 78:1-6.
12. Xia S. et al., 2020. The role of the furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Sig. Transduct. Ther. 5:92.
13. Korber B. et al., 2020.Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases the infectivity of the COVID-19 virus. Cell. 182:1-16.
14. Callaway E., 2020. Making sense of coronavirus mutations. Nature. 585:174-177.
15. Plante J.A. et al., 2020. Spike mutation D614G alters SARS-CoV-2 fitness and neutralization susceptibility. bioRxiv. doi: 10.1101/2020.09.01.278689.